 National Institutes of Health
National Institutes of HealthIn the race to develop a treatment for COVID-19, the disease threatening millions of lives around the world, scientists are studying every aspect of the SARS-CoV-2 coronavirus that causes it.
One vital step is understanding the precise shape and structure of the virus at the molecular level. Knowing the shape lets scientists identify targets and design drugs to hit them.
This approach was used to create the anti-influenza drugs oseltamivir and zanamivir (Tamiflu and Relenza) among others. It also shows great promise for the new coronavirus.
What is being done for COVID-19 treatment?
At present, scientists and clinicians are investigating the use of existing drugs such as chloroquine, which is used as an antimalarial and to treat autoimmune disorders, and the antiviral remdesvir. (Despite what you may read or hear, none of these are approved treatments for COVID-19 and should not be taken without medical instructions.)
Many pharmaceutical companies are also looking into therapies based on antibodies produced in people who have already been infected by the virus and recovered.
There are also several attempts in progress to create a vaccine which can be given to healthy people to make them immune. There are various approaches in play, including the use of genetic material or synthetic viral proteins to find ones that teach our immune systems to mount an effective defence against the virus.
Human trials are already under way for some of these experimental drugs and vaccines, and time will tell if they work. But almost certainly, an effective long-term strategy will also require significant research investment to allow the design of novel treatments that are specifically targeted for the SARS-CoV-2 virus.
How can you design new treatments without knowing what your target looks like?
Why structural biology?
If you want to design a key to a lock, it is much easier if you know what the lock looks like. In the same way, to design targeted treatments it is important to know what the target looks like.
In SARS-CoV-2 virus, the targets are its genetic material and proteins. These molecules let the virus invade humans and make multiple copies of itself.
Structural biology is a field of science that allows us to see beyond what is visible to the naked eye, to see nanoscopic-sized molecules such as DNA, RNA and proteins. Structural biology methods such as X-ray crystallography and cryo-electronmicroscopy are currently being used at a rapid pace to visualise molecular components of the virus.
Within weeks from when the genetic sequence of the SARS-CoV-2 was made available, structural biologists have used these techniques to see proteins that make up the SARS-CoV-2 virus.
Some of these include the “spike” protein, a protein that helps the virus to gain entry into the host and enzymes that enable virus replication including the main protease, an attractive target for drug development. The inner molecular machinery of the SARS-CoV-2 virus is just beginning to be revealed and there is more to come.
Read more: Revealed: the protein 'spike' that lets the 2019-nCoV coronavirus pierce and invade human cells
How is structural biology helping with COVID-19 research?
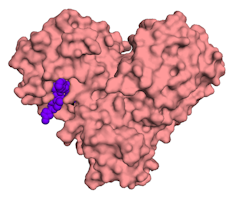 The main protease enzyme of the SARS-CoV-2 coronavirus, with a small chemical molecule in purple.Onisha Patel, Author provided
The main protease enzyme of the SARS-CoV-2 coronavirus, with a small chemical molecule in purple.Onisha Patel, Author providedScientists are using structural biology data to look for special features within the viral proteins. One such feature is a cavity or space where a small chemical molecule (a potential drug) can fit.
Once identified, researchers work on the molecule to improve the fit and make it work better as a drug. Eventually the chemical molecule might fit tightly enough to stop the viral protein from doing its job, much like a spanner thrown into a set of gears.
This approach to the design of new and targeted drugs is called “structure-based drug design”. It is more efficient, precise and time-saving. It has been used in the past to create anti-viral drugs such as oseltamivir (Tamiflu) and zanamivir (Relenza) that target the flu virus.
Scientists are now using this approach to discover drugs against SARS-CoV-2. For vaccine design and therapeutics development, knowing what the SARS-CoV-2 “spike” protein looks like is a major breakthrough. Scientists are already planning to use a stabilised version of this protein to screen for antibodies from people who have recovered from COVID-19 infection.
Where is structural biology data being shared?
Modern robotics, better instrumentation, faster data collection, better computing and software have revolutionised the speed with which structural biology information is being made available. This would not have been possible even five years ago.
The structural data collected is deposited at the Protein Data Bank (PDB) and the Electron Microscopy Data Bank (EMDB), online open access repositories who make this data available worldwide free of charge. This open access policy makes it possible for scientists globally to answer some of the most intriguing questions on SARS-CoV-2 biology and find the best ways to design new treatments.
Whether for new drugs or vaccines, structural biology is at the frontline to understand the molecular machinery from viruses to humans.
Onisha Patel receives funding from the National Health and Medical Research Council. She has previously received funding from the Australian Research Council.
Isabelle Lucet receives funding from the National Health and Medical Research. She has previously received funding from the Australian Research Council
Michael Roy receives funding from the National Health and Medical Research Council.
Authors: Onisha Patel, Senior Postdoctoral Research Fellow, Walter and Eliza Hall Institute
| < Prev | Next > |
|---|








